Overview
Our project aims to produce a pine species resistant to pine wilt disease caused by B. xylophilus. We use an RNA interference mechanism to solve this problem.
The key of our part design is to construct a self-complementary sequence with the ability to form a double-stranded RNA structure after transcription. This dsRNA targets genes of the B. xylophilus during the RNAi pathway.
Our parts are constructed into the T-DNA region of a binary vector, and the vector is transformed into Agrobacterium. The T-DNA is then transferred to the plant cell and integrated into the plant genome by Agrobacterium-mediated transformation.
As a result, the engineered plants can express the dsRNA. Once uptake by the nematode, the dsRNA carries out further RNAi-induced gene knockdown in the nematode, leading to loss of pathogenicity.
RNAi
As the weapon we use to tackle the problem, RNAi is a biological process in which RNA molecules inhibit gene expression or translation, mainly by degrading the mRNA molecules of target genes.
Generally for RNAi, the hairpin loop will be cleaved into dsRNA by Dicer, and if it is large enough, it’ll be cleaved into multiple 21bp fragments, then the sense strand is discarded. The assembly of the antisense strand with Ago/RISC complex and the activation of RISC will make it able to conduct target recognition, and finally cleavage the target mRNA or destabilize it.
The approach we chose to induce RNAi is the expression of long hairpin RNA (hpRNA). This method is facilitated by DNA constructs that enable insertion of ~400 bp complementary to our target gene. Since the expression of short hpRNA (shRNA), in which only a single ~21-nt interfering RNA is produced, might be less effective considering the possible mutations in our targets. We thought that multi-segment knockdown might be more effective, so we selected the former one.
Gene expressions of C. elegans can easily be controlled since they can feed on E. coli expressing dsRNA. However, B. xylophilus belongs to the stylet-type of nematode, so we chose pine callus to deliver dsRNA to B. xylophilus, which can be uptaken by it as the bait.
It may be worried that engineered parts in the callus genome may produce signal molecules that can move through the vascular system, leading to the specific silencing in certain plant genes in a local area or throughout the entire plant. However, in our case, it won’t happen because our target is the B. xylophilus gene. Moreover, because it does not affect the plant gametes it will not produce genetically modified plant offspring. The dsRNA will transfer from callus to B. xylophilus when they eat plant cells and then conduct the following knockdown of the target gene!

Target genes & plasmid
Choose the target gene
To prevent the nematode from causing pine wilt disease, the RNAi target gene must encode proteins that are essential to the pathogenicity of the nematode. Previous studies identified such specific proteins using gene silencing by dsRNA interference, providing a potential candidate as the parts of our project.
Bxpel1 (B. xylophilus pectate lyase 1)
B. xylophilus is a migratory nematode and it will propagate through the host tissue cell after the infection of pine wilt disease. The pectate lyase has been suggested as a cell wall–degrading enzymes involved in the interaction of the nematode and its host plant. As a result, silencing the pectate lyase gene by RNAi may reduce the migration speed of B. xylophilus.
Bx-Prx (B. xylophilus peroxiredoxin)
The peroxiredoxin is a ubiquitous thiol-specific peroxidase involved in the detoxification of cytotoxic peroxides. The major function of peroxiredoxin is to prevent cell damage caused by oxidative stress. Furthermore, Bx-Prx protein was proposed to be secreted from the body of B. xylophilus and may have served as an antioxidant for protecting it against host plant immune responses, such as oxygen free radicals in the pine tree. Relevant research shows that silencing of Bx-Prx gene by dsRNA decreased the pathogenicity of nematodes on pine tree seedlings.
However, the previous studies conducted the silencing experiment by RNA soaking method with high concentration of dsRNA. Since the goal of our project is to prevent pine trees from nematode-caused pine wilt disease, we try to construct a pine that can express the dsRNA of the target gene (Bxpel1 or Bx-PRx) with the ability to down regulate the gene expression in B. xylophilus after the infection. Loss of migration speed and pathogenicity in the nematodes may make them less harmful to pine trees.
Vector: pHANNIBAL
Usually, the transcription of target DNA sequence in a plasmid produces single-stranded RNA. However, RNA interference is a double-stranded RNA-mediated mechanism in cells, indicating that our part design must correspond to a vector which improves the conversion of the transcript into a double-stranded structure. pHANNIBAL is a vector used in previous research for the RNAi-mediated gene knockdown in plants. pHANNIBAL vector is reported to allow the insertion gene to be converted into a hairpin RNA (hpRNA) structure. because of the pdk intron within it. hpRNA is an RNA molecule with a hairpin loop and double-stranded self-complementary folding. The transcripted hpRNA could be then cut into siRNA by dicer, following the RNAi effect on target mRNA originally expressed in the cell.
Vector: pBI121
To conduct Agrobacterium-mediated transformation, we need the binary vector pBI121, which can carry out the Agrobacterium-mediated transformation on pine callus. The promoter of pBI121 is CaMV35S, which is a Pol II promoter and is widely used in transgenic plants, and hence helpful for the transformation. However, pBI121 lacks the necessary element to form a double-stranded transcript. Thus, we insert the pdk intron taken out from pHANNIBAL into pBI121, so we can get a vector with both the ability to transform plant cells and perform RNAi.
How our parts work
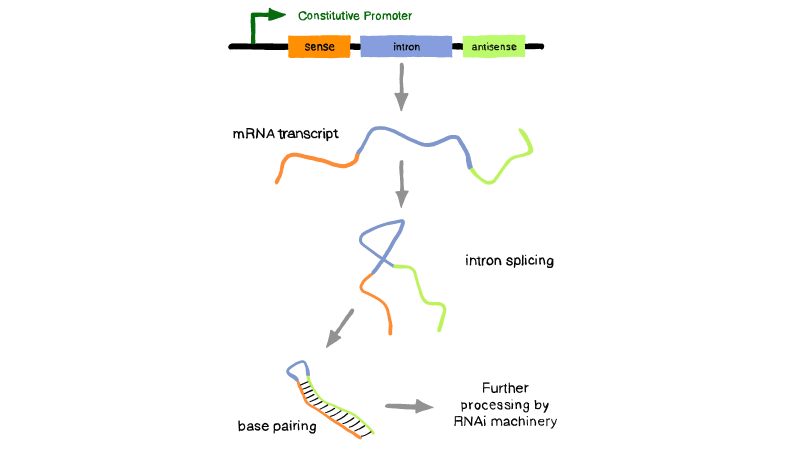
For our parts, the simple explanation is shown below. A general lhpRNA design contains a sense fragment, an antisense fragment, and an intron. After transcription and intron splicing, it’ll turn into the double-stranded structure for further process of RNAi. Among our parts, we choose pdk intron, which is native to our plasmid- pHANNIBAL. After two rounds of transformation, it will be inserted into pHANNIBAL. (First time the sense fragment and then the antisense fragment.) To make our sequence more consistent with the target B. xylophilus gene, we chose the exon fragment close to the intron in the target B. xylophilus gene as our sense and antisense strand.
And if we want to use pHANNIBAL as our vector, we need to conduct two rounds of transformation in order to make both sense and antisense strands inside the vector. Therefore, we also designed another sequence that contains sense, antisense strands, and the intron. It’ll be transformed into another plasmid, pBI121. And this time, we only need one round of transformation.
Let’s talk about what’ll happen to our target genes- Bx-Prx, Bxpel1, and BxGPCR. Bx-Prx is associated with the reproduction and pathogenicity of B. xylophilus. This gene encodes peroxiredoxin, a protein that influences thioredoxin peroxidase activity. Peroxiredoxin might play a role in protecting B. xylophilus from oxygen free radicals in the pine tree. As for Bxpel1, it encodes pectate lyase.
As we mentioned above, after our plasmid is transferred to B. xylophilus, it has been cleaved into many 21bp mRNAs by Dicer and will bind to the complementary mRNA produced in it, then degrade the target gene. Take the expression of Bx-prx as an example:
(1) In the case that Bx-prx expressed
Prx could be secreted to neutralize H2O2 produced by the host.
-->The neutralization and the following inhibition of the SA accumulation
-->The retard of systematic SR reactions in hosts.
(2) If knockdown by RNAi
Infestation of B. xylophilus induces H2O2 defense in pine.
-->Increase of SA in host plants.
-->Increase the expression of pathogenesis-related protein(PRP) and signal molecular (SM) lead to the following systemic resistance (SR) reactions in pines.
Deliver from Agrobacterium to callus
To deliver our part into plant callus, we use Agrobacterium tumefaciens LBA4404 strain. Agrobacterium is a parasitic bacteria that grow on plants. The bacterial infection of Agrobacterium is caused by integrating transfer DNA (T-DNA) into the host plant genome. This Agrobacterium-mediated plant transformation mechanism involves several essential virulence genes that are necessary to transfer T-DNA from Agrobacterium to the plant nucleus. Therefore, we use the binary vector system as a method to perform plant transformation.
A. tumefaciens LBA4404 strain is optimized for the transformation efficiencies. It contains the first vector in the binary system, the Ti plasmid pAL4404, which has the vir gene encoding bacterial protein machinery. These vir proteins are involved in the T-DNA processing. However, the Ti plasmid is usually large in size and has a low copy number, which makes it difficult for the cloning process. To solve this problem, the T-DNA region is carried by another vector, the pBI121 binary vector, with a relatively high copy number. Most importantly, the binary vector includes the left and right T-DNA borders sequence defining the T-DNA region. In our project, our engineered parts are designed to insert into the T-DNA region of the binary vector and will be integrated into the plant genome during Agrobacterium-mediated transformation.
Reference
[1] Silva, D.; Jemc, J. C., Sorting Out Identities: An Educational Primer for Use with “Novel Tools for Genetic Manipulation of Follicle Stem Cells in the Drosophila Ovary Reveal an Integrin-Dependent Transition from Quiescence to Proliferation”. Genetics 2015, 201 (1), 13-22.
[2] Wilson, R. C.; Doudna, J. A., Molecular mechanisms of RNA interference. Annual review of biophysics 2013, 42, 217-239.
[3] Dunoyer, P.; Himber, C.; Ruiz-Ferrer, V.; Alioua, A.; Voinnet, O., Intra-and intercellular RNA interference in Arabidopsis thaliana requires components of the microRNA and heterochromatic silencing pathways. Nature genetics 2007, 39 (7), 848-856.
[4] Qiu, X.-W.; Wu, X.-Q.; Huang, L.; Ye, J.-R. Influence of Bxpel1 Gene Silencing by DsRNA Interference on the Development and Pathogenicity of the Pine Wood Nematode, Bursaphelenchus Xylophilus. Int. J. Mol. Sci. 2016, 17 (1), 125.
[5] Kikuchi, T.; Shibuya, H.; Aikawa, T.; Jones, J. T. Cloning and Characterization of Pectate Lyases Expressed in the Esophageal Gland of the Pine Wood Nematode Bursaphelenchus Xylophilus. Mol. Plant. Microbe. Interact. 2006, 19 (3), 280–287.
[6] Fu, H.-Y.; Ren, J.-H.; Huang, L.; Li, H.; Ye, J.-R. Screening and Functional Analysis of the Peroxiredoxin Specifically Expressed in Bursaphelenchus Xylophilus--the Causative Agent of Pine Wilt Disease. Int. J. Mol. Sci. 2014, 15 (6), 10215–10232.
[7] Bakhetia, M.; Charlton, W. L.; Urwin, P. E.; McPherson, M. J.; Atkinson, H. J. RNA Interference and Plant Parasitic Nematodes. Trends Plant Sci. 2005, 10 (8), 362–367.
[8] Wesley, S. V.; Helliwell, C. A.; Smith, N. A.; Wang, M. B.; Rouse, D. T.; Liu, Q.; Gooding, P. S.; Singh, S. P.; Abbott, D.; Stoutjesdijk, P. A.; Robinson, S. P.; Gleave, A. P.; Green, A. G.; Waterhouse, P. M. Construct Design for Efficient, Effective and High-Throughput Gene Silencing in Plants: Construct Design for Gene Silencing. Plant J. 2001, 27 (6), 581–590.
[9] Wesley, S. V.; Helliwell, C.; Wang, M.-B.; Waterhouse, P. Posttranscriptional Gene Silencing in Plants. Methods Mol. Biol. 2004, 265, 117–129.
[10] Wesley, S. V.; Helliwell, C. A.; Smith, N. A.; Wang, M.; Rouse, D. T.; Liu, Q.; Gooding, P. S.; Singh, S. P.; Abbott, D.; Stoutjesdijk, P. A., Construct design for efficient, effective and high‐throughput gene silencing in plants. The Plant Journal 2001, 27 (6), 581-590.
[11] Fu, H.-Y.; Ren, J.-H.; Huang, L.; Li, H.; Ye, J.-R., Screening and functional analysis of the peroxiredoxin specifically expressed in Bursaphelenchus xylophilus—the causative agent of pine wilt disease. International journal of molecular sciences 2014, 15 (6), 10215-10232.
[12] Qiu, X.-W.; Wu, X.-Q.; Huang, L.; Ye, J.-R., Influence of bxpel1 gene silencing by dsRNA interference on the development and pathogenicity of the pine wood nematode, Bursaphelenchus xylophilus. International journal of molecular sciences 2016, 17 (1), 125.
[13] Li, Z.; Zhang, Q.; Zhou, X., A 2-Cys peroxiredoxin in response to oxidative stress in the pine wood nematode, Bursaphelenchus xylophilus. Scientific reports 2016, 6 (1), 1-10.
[14] Hoekema, A.; Hirsch, P. R.; Hooykaas, P. J. J.; Schilperoort, R. A. A Binary Plant Vector Strategy Based on Separation of Vir- and T-Region of the Agrobacterium Tumefaciens Ti-Plasmid. Nature 1983, 303 (5913), 179–180.

